Meet PepCNN: A Deep Learning Tool for Predicting Peptide Binding Residues in Proteins Using Sequence, Structural, and Language Model Features
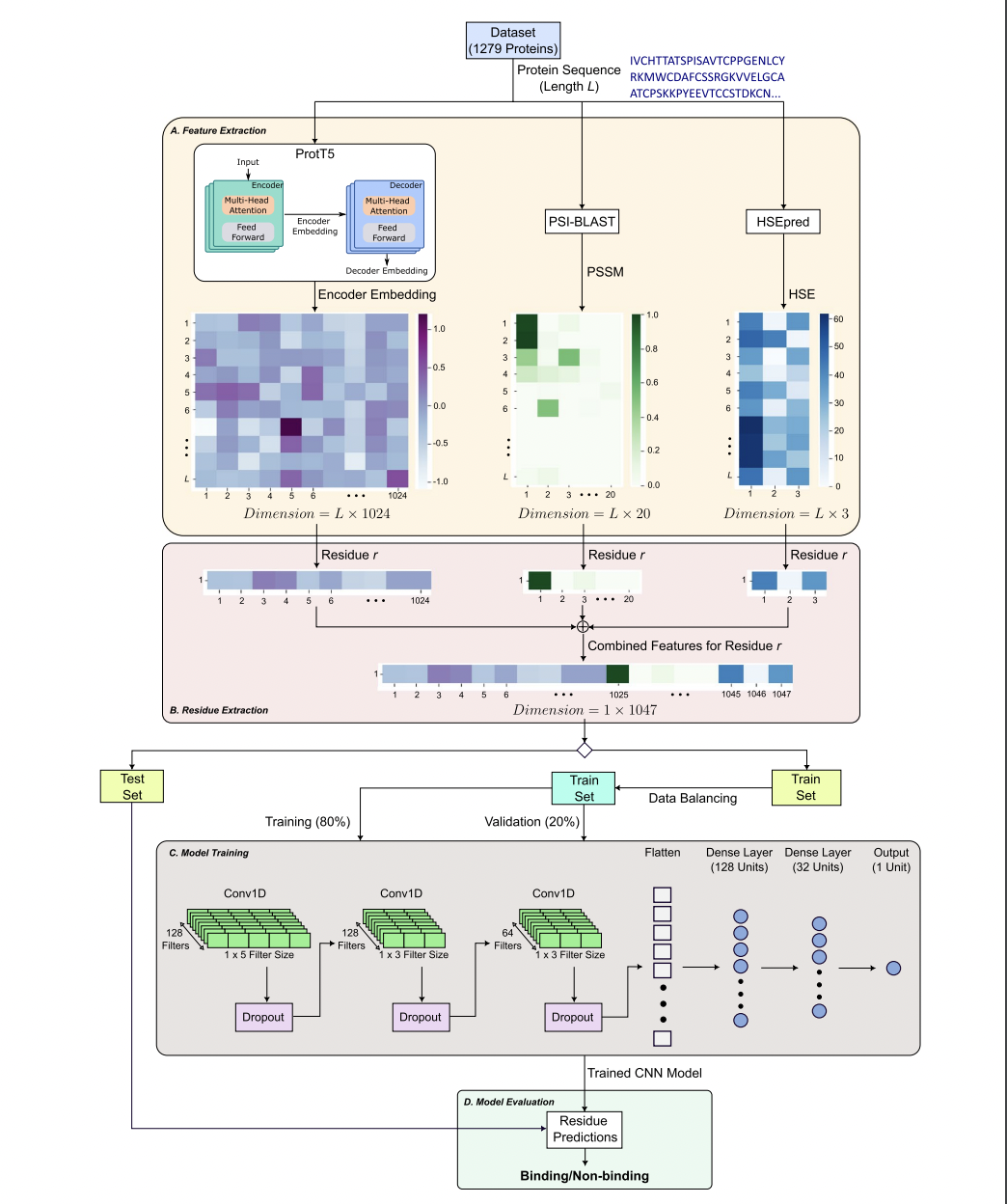
PepCNN, a deep learning model developed by researchers from Griffith University, RIKEN Center for Integrative Medical Sciences, Rutgers University, and The University of Tokyo, addresses the problem of predicting protein-peptide binding residues. PepCNN outperforms other methods in terms of specificity, precision, and AUC metrics by combining structural and sequence-based information, making it a valuable tool for understanding protein-peptide interactions and advancing drug discovery efforts.
Understanding protein-peptide interactions is crucial for cellular processes and disease mechanisms like cancer, necessitating computational methods as experimental approaches are resource-intensive. Computational models, categorized into structure-based and sequence-based, offer alternatives. Utilizing features from pre-trained protein language models and exposure data, PepCNN outperforms previous methods, emphasizing the significance of its feature set for improved prediction accuracy in protein-peptide interactions.
There is a need for computational approaches to gain a deeper understanding of protein-peptide interactions and their role in cellular processes and disease mechanisms. While structure-based and sequence-based models have been developed, accuracy remains a challenge due to the complexity of the interactions. PepCNN, a novel deep learning model, has been proposed to solve this challenge by integrating structural and sequence-based information to predict peptide binding residues. With superior performance compared to existing methods, PepCNN is a promising tool for supporting drug discovery efforts and advancing the understanding of protein-peptide interactions.
PepCNN utilizes innovative techniques such as half-sphere exposure, position-specific scoring matrices, and embedding from a pre-trained protein language model to achieve superior results compared to nine existing methods, including PepBCL. Its impressive specificity and precision stand out, and its performance surpasses other state-of-the-art methods. These advancements highlight the effectiveness of the proposed method.
The deep learning prediction model, PepCNN, outperformed various methods, including PepBCL, with higher specificity, precision, and AUC. After being evaluated on two test sets, PepCNN displayed notable improvements, particularly in AUC. The results showed that sensitivity was 0.254, specificity was 0.988, precision was 0.55, MCC was 0.350, and AUC was 0.843 on the first test set. Future research aims to integrate DeepInsight technology to facilitate the application of 2D CNN architectures and transfer learning techniques for further advancements.
In conclusion, the advanced deep-learning prediction model, PepCNN, incorporating structural and sequence-based information from primary protein sequences, outperforms existing methods in specificity, precision, and AUC, as demonstrated in tests conducted on TE125 and TE639 datasets. Further research aims to enhance its performance by integrating DeepInsight technology, enabling the application of 2D CNN architectures and transfer learning techniques.
Check out the Paper and Github. All credit for this research goes to the researchers of this project. Also, don’t forget to join our 33k+ ML SubReddit, 41k+ Facebook Community, Discord Channel, and Email Newsletter, where we share the latest AI research news, cool AI projects, and more.
If you like our work, you will love our newsletter..
Sana Hassan, a consulting intern at Marktechpost and dual-degree student at IIT Madras, is passionate about applying technology and AI to address real-world challenges. With a keen interest in solving practical problems, he brings a fresh perspective to the intersection of AI and real-life solutions.

